Crossover (genetic algorithm)
In genetic algorithms, crossover is a genetic operator used to vary the programming of a chromosome or chromosomes from one generation to the next. It is analogous to reproduction and biological crossover, upon which genetic algorithms are based. Cross over is a process of taking more than one parent solutions and producing a child solution from them. There are methods for selection of the chromosomes.Those are also given below.
Contents |
Methods of selection of chromosomes for crossover
- Roulette wheel selection
It is also known as fitness proportionate selection.The individual is selected on the basis of fitness.The probability of an individual to be selected increases with the fitness of the individual greater or less than its competitor's fitness.
- Boltzmann selection
- Tournament selection
- Rank selection
- Steady state selection
Crossover techniques
Many crossover techniques exist for organisms which use different data structures to store themselves.
One-point crossover
A single crossover point on both parents' organism strings is selected. All data beyond that point in either organism string is swapped between the two parent organisms. The resulting organisms are the children:
Two-point crossover
Two-point crossover calls for two points to be selected on the parent organism strings. Everything between the two points is swapped between the parent organisms, rendering two child organisms:
"Cut and splice"
Another crossover variant, the "cut and splice" approach, results in a change in length of the children strings. The reason for this difference is that each parent string has a separate choice of crossover point.
Uniform Crossover and Half Uniform Crossover
The Uniform Crossover uses a fixed mixing ratio between two parents. Unlike one- and two-point crossover, the Uniform Crossover enables the parent chromosomes to contribute the gene level rather than the segment level. If the mixing ratio is 0.5, the offspring has approximately half of the genes from first parent and the other half from second parent, although cross over points can be randomly chosen as seen below
The Uniform Crossover evaluates each bit in the parent strings for exchange with a probability of 0.5. Even though the uniform crossover is a poor method, empirical evidence suggest that it is a more exploratory approach to crossover than the traditional exploitative approach that maintains longer schemata. This results in a more complete search of the design space with maintaining the exchange of good information. Unfortunately, no satisfactory theory exists to explain the discrepancies between the Uniform Crossover and the traditional approaches. [1]
In the uniform crossover scheme (UX) individual bits in the string are compared between two parents. The bits are swapped with a fixed probability, typically 0.2.
In the half uniform crossover scheme (HUX), exactly half of the nonmatching bits are swapped. Thus first the Hamming distance (the number of differing bits) is calculated. This number is divided by two. The resulting number is how many of the bits that do not match between the two parents will be swapped.
Three parent crossover
In this technique,the child is derived from three parents.They are randomly chosen.Each bit of first parent is checked with bit of second parent whether they are same.If same then the bit is taken for the offspring otherwise the bit from the third parent is taken for the offspring.
parent1 1 1 0 1 0 0 0 1 0
parent2 0 1 1 0 0 1 0 0 1
parent3 1 1 0 1 1 0 1 0 1
offspring 1 1 0 1 0 0 0 0 1[2]
Crossover for Ordered Chromosomes
Depending on how the chromosome represents the solution, a direct swap may not be possible.
One such case is when the chromosome is an ordered list, such as an ordered list of the cities to be travelled for the traveling salesman problem. There are many crossover methods for ordered chromosomes. The already mentioned N-point crossover can be applied for ordered chromosomes also, but this always need a corresponding repair process, actually, some ordered crossover menthod are derived from the idea. However, sometimes a crossover of chromosomes produces recombinations which violate the constraint of ordering and thus need to be repaired. Several examples for crossover operators (also mutation operator) preserving a given order are given in [3]:
- partially-matched crossover (PMX): In this method, two crossover points are selected at random and PMX proceeds by position wise exchanges.The two crossover points give matching selection.It affects cross by position-by-position exchange operations.In this method parents are mapped to each other hence we can also call it partially mapped crossover.[4]
- cycle crossover (CX): Beginning at any gene
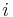 in parent 1, the
in parent 1, the 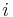 -th gene in parent 2 becomes replaced by it. The same is repeated for the displaced gene until the gene which is equal to the first inserted gene becomes replaced (cycle).
-th gene in parent 2 becomes replaced by it. The same is repeated for the displaced gene until the gene which is equal to the first inserted gene becomes replaced (cycle). - order crossover operator (OX1): A portion of one parent is mapped to a portion of the other parent. From the replaced portion on, the rest is filled up by the remaining genes, where already present genes are omitted and the order is preserved.
- order-based crossover operator (OX2)
- position-based crossover operator (POS)
- voting recombination crossover operator (VR)
- alternating-position crossover operator (AP)
- sequential constrictive crossover operator (SCX) [5]
Other possible methods include the edge recombination operator and partially mapped crossover.
Crossover biases
For crossover operators which exchange contiguous sections of the chromosomes (e.g. k-point) the ordering of the variables may become important. This is particularly true when good solutions contain building blocks which might be disrupted by a non-respectful crossover operator.
See also
References
- John Holland, Adaptation in Natural and Artificial Systems, University of Michigan Press, Ann Arbor, Michigan. 1975. ISBN 0-262-58111-6.
- Larry J. Eshelman, The CHC Adaptive Search Algorithm: How to Have Safe Search When Engaging in Nontraditional Genetic Recombination, in Gregory J. E. Rawlins editor, Proceedings of the First Workshop on Foundations of Genetic Algorithms. pages 265-283. Morgan Kaufmann, 1991. ISBN 1-55860-170-8.
- Tomasz D. Gwiazda, Genetic Algorithms Reference Vol.1 Crossover for single-objective numerical optimization problems, Tomasz Gwiazda, Lomianki, 2006. ISBN 83-923958-3-2.
- ^ (eds.), P.K. Chawdhry ... (1998). Soft computing in engineering design and manufacturing. London: Springer. pp. 164. ISBN 3540762140. http://books.google.com/books?id=mxcP1mSjOlsC.
- ^ Introduction to genetic algorithms By S. N. Sivanandam, S. N. Deepa
- ^ Pedro Larrañaga et al., "Learning Bayesian Network Structures by searching for the best ordering with genetic algorithms", IEEE Transactions on systems, man and cybernetics, Vol 26, No. 4, 1996
- ^ Introduction to genetic algorithms By S. N. Sivanandam, S. N. Deepa
- ^ Ahmed, Zakir H. "Genetic Algorithm for the Traveling Salesman Problem Using Sequential Constructive Crossover Operator." International Journal of Biometric and Bioinformatics 3.6 (2010). Computer Science Journals. Web. <http://www.cscjournals.org/csc/manuscript/Journals/IJBB/volume3/Issue6/IJBB-41.pdf>.
External links
- Newsgroup: comp.ai.genetic FAQ - see section on crossover (also known as recombination).